As successful as the COVID vaccines have been, many experts see inoculations based on natural proteins as an interim technology. “It’s becoming clear that just delivering natural or stabilized proteins is not sufficient,” says Rino Rappuoli, chief scientist and head of vaccine development at U.K.-based pharmaceutical giant GlaxoSmithKline. Most current vaccines, from childhood inoculations to adult flu shots, involve such natural proteins, which vaccinologists call immunogens; GSK makes a lot of them. “We need to design immunogens that are better than natural molecules,” Rappuoli says.
Walls and Veesler had an idea. What if, instead of a whole spike, the immune system were presented with just the RBD tip, which would not have any shield to hide behind? “We wanted to put the key component on display,” Walls says, “to say, ‘Hey, immune system, this is where you want to react!’
The immediate trouble with that notion was that biology does not make isolated RBDs, and the segment on its own would be too small and unfamiliar to get the immune system’s attention. But Walls and Veesler knew some people who could help them solve that problem. Just up the street from them was the Bell Labs of protein invention, the University of Washington’s Institute for Protein Design (IPD). The institute had learned enough about protein folding to design and build a few hundred very simple, small proteins—unlike any that have ever been found in a living organism—that would fold into consistent shapes with predictable functions.
In 2019 a group in the IPD led by biochemist Neil King had designed two tiny proteins with complementary interfaces that, when mixed together in solution, would snap together and self-assemble into nanoparticles. These balls were about the size of a virus and were completely customizable through a simple change to their genetic code. When the scientists festooned the particles with 20 protein spikes from the respiratory syncytial virus, the second-leading cause of infant mortality worldwide, they triggered an impressive immune response in early tests.
Why not try a similar nanoparticle core for a SARS-CoV-2 vaccine, Walls and Veesler thought, using just the RBD instead of an entire spike? As a bonus, the protein-based nanoparticle would be cheap and fast to produce compared with vaccines that use killed or weakened virus. It would also be stable at room temperature and easy to deliver to people, unlike fragile mRNA vaccines that must be kept in a deep freeze.
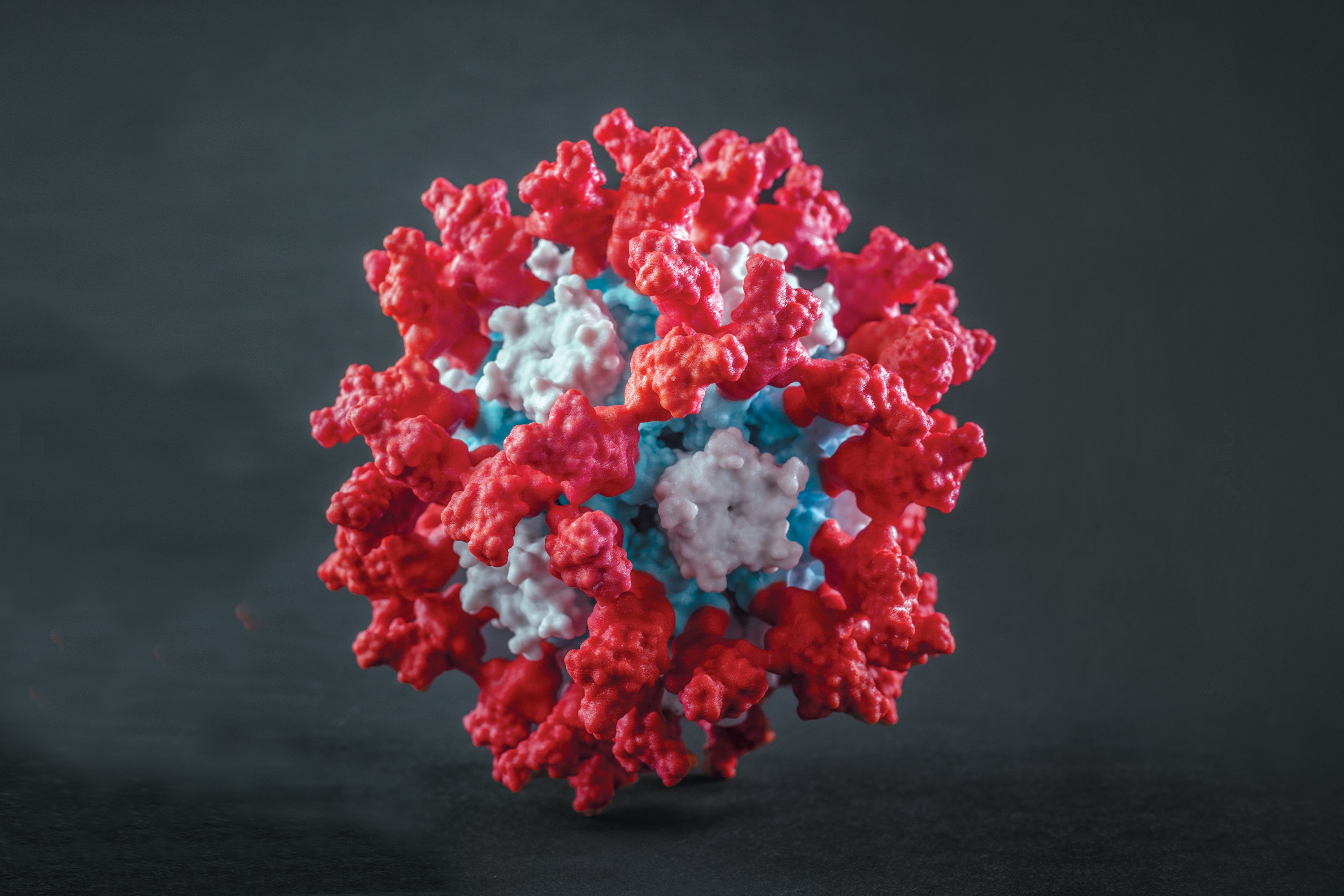
Walls reached out to the IPD and collaborated with nanoparticle specialist Brooke Fiala, who worked with King, on a prototype—a nanoparticle sphere displaying 60 copies of the RBD. The scientists also tried something radical: Instead of fusing the RBDs directly to the surface of the nanoparticle, they tethered them with short strings of amino acids, like kites. Giving the RBDs a little bit of play could allow the immune system to get a better look at every angle and produce antibodies that would attack many different spots.
But nobody knew whether that would really happen. So on that April Friday last year, as Walls waited for results, she had her fingers crossed. Three weeks earlier she and her colleagues had injected some mice with the nanoparticle vaccine. Other mice got the plain spike that other vaccines were using. Now the researchers had drawn blood from the mice and mixed it with a SARS-CoV-2 pseudovirus, an artificial, nonreplicating version of the virus that is safer to use in labs. The idea was to see whether any vaccinated mice had developed antibodies that would home in on and neutralize the pseudovirus.
It takes a while for antibodies to do their thing, which is why Walls had to wait until late that Friday night. No way was she going home to be kept in suspense all weekend. Her colleagues had wished her good luck as they headed out the door. Before Veesler cut out, he asked her to contact him as soon as she had results.
Now it was dark outside, and the lab was ghostly quiet. It was finally time to look. Walls fired up a lab instrument that could detect and count antibodies attached to virus particles, took a deep breath and peeked at the numbers.
Some mice had been given a low dose of the plain spike, and that was a total failure: zero effect on the pseudoviruses. Mice given a high dose of the spike showed antibodies with a moderate neutralizing effect, similar to what some other vaccines had produced. But in mice that got the nanoparticle vaccine, the pseudovirus was completely outmatched. Antibodies smothered it and had 10 times the neutralizing effect of the large-dose spike preparation. That magnitude held even when only a minuscule dose was used. Walls was looking at something that could be a low-cost, shelf-stable, ultrapotent vaccine.
Walls fired off an all-caps text message to Veesler: “THEY’RE NEUTRALIZING!”
Veesler wrote right back: “The next generation of coronavirus vaccines is in your hands!”
That was only the first of several tests the vaccine had to pass. From there they would have to prove the vaccine could offer protection from the live virus in mice, nonhuman primates and, finally, people. The nanoparticles entered that last testing phase early in 2021. But at that moment, as an emblem of the power of protein design, it was already a success—the clearest sign yet that a technology long beyond our grasp had suddenly arrived. We were learning to sculpt the living clay from which we are all made.
As transformational as the genetics revolution of the past decades has been, at its heart has always been a mystery: proteins. A gene is simply the code for making a single protein. In that gene, a set of three DNA nucleotides, represented by letters, yields one amino acid, and another triplet codes for a different amino acid. There are 20 amino acids that a cell can use as protein-building blocks, and each one has a unique shape and function. Some are more flexible than others. Some are positively charged, some negative. Some are attracted to water; others are repelled by it.
All day long our cells churn out new proteins in the exact order of amino acids dictated by our genetic code, and the proteins spontaneously snap into shape. That shape, along with the charges of the atoms on the exposed bits, determines the function: what they respond to, what they attach to, what they can do. When we say, “He has the gene for red hair,” it means he has the blueprint for proteins that lead to a particular kind of pigment. When we say, “She has a gene that causes breast cancer,” it means she has a mutation in a gene that causes its protein to be made with an incorrect amino acid, which screws up its function in a way that can lead to cancer.
Understanding the mechanics of protein folding would allow us to design new classes of drugs that could hobble or replace proteins gone wrong and to probe the etiology of diseases such as Alzheimer’s, Parkinson’s, Huntington’s and cystic fibrosis, which are linked to misshapen proteins.
Unfortunately, because proteins are so small, it is almost impossible to tell what is happening in this nanoworld, even with powerful microscopes. We do not know precisely how all of these proteins fold correctly, much less what goes wrong when they misfold. It can take a year and $120,000 to produce a high-resolution image of one protein on specialized equipment. We currently know the structures of just 0.1 percent of them. For the rest, we guess. That is why there is a mystery at the center of the genetics revolution: Certain genetic sequences are associated with physical and mental effects, but often we cannot tell why. We have lacked the Rosetta stone of protein structure to translate between the starting point of genes and the end point of bodily functions.
In theory, it should be possible to predict the final structure of a protein from its genetic sequence—a task so essential to our understanding that in 2005 Science magazine included it in its 125th-anniversary issue’s list of the most important unanswered questions in science. But in reality, it has been possible for only a very few extremely simple proteins. For example, scientists know that if they want to build a straight helix (a common Slinky-like structure in proteins that provides stability), they can use amino acids such as leucine, alanine and glutamate, which have the right curve and complementarity to form regular spirals and bond tightly to the amino acids on the coil above or below them. If scientists want a kink in their Slinky, they can add a proline, which does not form a bond and allows the rest of the helix to bend away from it.
Structural biologists such as David Baker, who founded the IPD—where Walls and Veesler went to get their nanoparticles—have been able to deduce a few of these basic rules. Baker’s group has incorporated these rubrics into a structure-predicting computer program called Rosetta and used them to make a number of small proteins, typically a few dozen amino acids in size. Some of their successes have shown the great potential of the field: microscopic “nanocages” that could be used to package drugs and transport them into the body and molecular detectors that go off when they encounter cells with specific combinations of amino acids on their surface, indicating such cells are cancerous.
But most important proteins in living things are much bigger than these examples and contain thousands of amino acids, each of which interacts with up to a dozen neighbors, some forming bonds as strong as those in a diamond, some pushing others away. All those relationships morph depending on proximity. So the possibilities quickly become astronomical, and the formulas for figuring out the final structures have long eluded our best minds and supercomputers.
Frustrated by this problem back in 1994, a group of computational biologists decided that a little friendly competition might spur some progress. Led by John Moult of the University of Maryland, they launched CASP, the Critical Assessment of Structure Prediction contest. Moult obtained detailed specs of proteins whose structure had been recently identified but not released. He sent the genetic sequence for the proteins to various teams from different research labs, which then submitted their best ideas about what the finished protein looked like.
Those predictions were scored on their similarity to the actual structure based on the percentage of molecules in the right place. Getting the basic architecture right might score a 50, getting the angles and links between the main parts might be good for a 70, and nailing the tiny molecular threads that sprout off proteins like hairs would merit a 90-plus.
Moult has been running the contest every two years since then. For a long time not even the best teams could do much better than guesswork. In 2012, the year Baker’s protein design institute started up, the very best CASP teams were averaging scores in the low 20s, and there had been no improvement for a decade. “There were moments after some CASPs where I’d see the results and despair,” Moult says. “I’d think, ‘This is all a joke. Why are we even doing this?’” Some new insights led to a rise at CASP11, with the best scores averaging nearly 30, and another slight bump to around 40 at CASP12.
Then came CASP13 in 2018. The best teams, led by Baker’s institute, improved again, averaging nearly 50, but they were bested by a surprise entrant: Google’s DeepMind, whose artificial-intelligence system had trounced the world’s best Go player in 2017. The AI averaged a score of about 57 per protein.
That result rocked the world’s protein-engineering labs, but it turned out to be just a dress rehearsal for 2020. In that year DeepMind’s predictions were spot-on. “I thought, ‘This can’t be right. Let’s wait for the next one,’” Moult says. “And they just kept coming.”
DeepMind averaged a 92 for all proteins. On the easier ones, it had virtually every atom in the right place. But its most impressive results were on some exceedingly difficult proteins that completely stymied most teams. On one molecule, no group scored higher than the 20s—DeepMind scored in the high 80s.
Moult was stunned by the results. “I spent a lot of my career on this,” he says. “I never thought we’d get this level of atomic accuracy.” Most impressive, he says, is the indication that DeepMind has picked up on previously unknown fundamentals. “It’s not just pattern recognition. In some alien way, the machine ‘understands’ the physics and can calculate how the atoms in a unique arrangement of amino acids are going to arrange themselves.”
“It was shocking,” agrees structural biologist and CASP competitor Mohammed AlQuraishi of Columbia University. “Never in my life had I expected to see a scientific advance so rapid.” AlQuraishi expects the breakthrough to transform the biological sciences.
The DeepMind team is expected to publish its methods paper, with details about how it worked, later this year. Some aspects may remain inscrutable—the AI picks up on faint relationships that cannot easily be explained with rules—but at the moment, scientists do have the general outlines.
To predict amino acids’ effects on one another, the machine’s programmers invoked a technique called attention that has been responsible for recent leaps forward in accurate language translation by AIs. Like proteins, language is a seemingly linear string of information that folds back on itself to produce meaning. A word such as “it” might draw its significance from a word used in an entirely different sentence. (“For the longest time, AI made no sense to me. And then, after much reading, I finally understood it.”) When we communicate, we are constantly moving backward and forward along this linear string, paying attention to one local cluster of words to understand what a different word means in context. Once we have that meaning resolved, we can move to another, related passage and understand those words in light of the new information.
DeepMind does something like this for proteins, focusing its attention on one local cluster of amino acids, understanding as much as it can about their relation to one another. Some pairs of aminos, for example, appear to have coevolved, indicating a bond between them and limiting their possible positions in the protein. DeepMind uses this information to leap to a different part of the protein and analyze that section in light of what it knows about the first cluster. It carries out multiple iterations across all parts of the protein string and eventually uses this information to build a 3-D cloud of points that represents the relations among all the atomic constituents of every amino. It basically treats protein folding like a new, alien language to be deciphered.
As other labs incorporate DeepMind’s techniques and on-point protein prediction becomes ubiquitous, AlQuraishi says, the lengthy trial-and-error period of getting a real-world protein to fold like you thought it would will become much faster. “It will percolate everywhere,” he says. “It’s going to make protein design much more effective.”

But the DeepMind team is not in the business of applied science, so the AI will not spend its time churning out blueprints for complicated protein construction on demand. Its big contribution will be indirect. “Their work shines light on the power of proteins and the bright future of engineering new ones,” says California Institute of Technology biochemist Frances Arnold, who won the Nobel Prize in Chemistry in 2018 for improving the performance of natural proteins through a method called directed evolution. “But they have not solved the problem of designing or engineering proteins to solve problems for people.”
That work will fall to the Arnolds and Bakers of the world, who are trying to use DeepMind’s techniques to supercharge their labs’ abilities to sculpt proteins. “It’s a big breakthrough,” says Baker, whose team again finished a distant second in the competition. “I think it will make what’s already working well work even better.”
Right now there is an enormous problem for people, to use Arnold’s phrase, that is wracking the world. That problem is COVID. When it hit, Baker and others in his lab looked to proteins for solutions. They plugged the genetic sequence for the coronavirus into Rosetta, their protein-structure-prediction computer program, to produce a 3-D model, then pored over it for weaknesses like Rebel pilots plotting an assault on the Death Star. As Walls did, they zeroed in on the spike’s RBD. But instead of making a vaccine to trigger antibody production, Baker wanted to build a better antibody. He wanted a protein whose sole purpose was to ensnare the RBD like microscopic Velcro.
Amazing as they are, antibodies are not perfect. The body cannot custom-design an antibody in advance for a pathogen it has never seen, so it makes a lot of different versions. When a new invader shows up, immune system cells make many copies of whatever antibody binds best, but the fit is not always tight enough to stop the pathogen. Natural antibodies are also relatively big proteins that are not always able to get their business end snug against a virus’s RBD.
Enter the “mini binders,” as Baker calls them. These are small synthetic proteins that can be designed amino acid by amino acid to fit precisely against a virus’s RBD. With no extraneous bits, they bind more tightly. And they are small and lightweight enough to be administered through a spritz up the nose rather than an injection into the arm. No needles!
Baker’s dream was to create a medication rather than a vaccine: a nasal spray that could be used at the first sign of infection—or beforehand as daily prevention—to flood the nose with a mist of mini binders that would coat the RBDs of virus particles before they could attach to anything. It would have the long shelf life of a bag of dried lentils, and it could be quickly reformulated for any new pathogen and rushed into the hands of health-care workers, teachers and anyone else on the front lines—a kind of designer-driven immune system for civilization.
To engineer the mini binder, Longxing Cao, a postdoc in Baker’s lab who headed the project, scouted the virus RBD’s structure, comparing it with the library of tiny proteins the institute had previously designed and looking for complementary shapes. Like a rock climber on a challenging face, the mini binder needed to be small enough to wriggle into the cleft where the RBD lay, and it needed to be shaped so that it could get firm handholds and toeholds in the right places. Cao cataloged where the RBD’s amino acids made patches of positive electrical charges, patches of negative charges and hydrophobic (water-hating) patches, then tailored mini binders to have as many complementary patches as possible. He tested millions of possibilities on Rosetta.
The best designs were made of three helixes connected like sausage links by short strings of amino acids. Each mini binder was about 60 amino acids long in total—less than a tenth the size of an antibody and a twentieth the size of a coronavirus spike.
Then, of course, Cao had to take his protein from Rosetta to the real world. Amazingly, that process has become trivially easy. DNA—the As, Cs, Gs and Ts of the genetic code—can be printed for pennies on devices that resemble inkjet printers. Cao printed DNA strands with the sequence for his mini binder and inserted them into yeast, which, like programmable livestock, pumped out those tiny proteins along with their normal ones. He then harvested the proteins and tested them.
The top mini binder bound the virus six times more effectively than the best antibodies known—better than any molecule on the planet, in fact, forming dozens of strong bonds with the RBD. It was extraordinarily stable, and it sprayed easily out of a nozzle. Hamsters given a snootful became immune to COVID. “I was definitely excited,” Cao says, “but not totally surprised.” Researchers expect clinical trials for mini binders to start later in 2021, and a number of labs around the world are now exploring other ways that mini proteins might help the body function or ward off illness.
Although there is great optimism about the technology, some biosecurity researchers have expressed concerns about proteins that could be designed for nefarious purposes. Prions, for example, which are responsible for “mad cow” and other neurodegenerative diseases, are misfolded proteins that cause other proteins to misfold in turn, triggering deadly chain reactions that are transmissible; they could be delivered by aerosol. The Biological Weapons Convention, which virtually all nations have signed, effectively bans the development or use of pathogen-based bioweapons, but no one ever thought to extend it to address proteins that were never part of an organism.
“This is a real concern,” says biosecurity expert Filippa Lentzos of King’s College London, “because potential future biological weapons won’t necessarily make us sick using pathogens.” Synthetic mini proteins may or may not fall under the control of the convention, she says, “so legal status is an important issue.”
But engineered mini proteins are also an extremely unlikely threat, Lentzos says, and quite low on her list of worries: “If you want to cause harm, why would you turn to something as sophisticated and complicated as protein design? There are plenty of more accessible things in nature you could use.” Naturally occurring toxins and pathogens are ready-made and all over the place. If you really want to hurt people, there are easier ways.
At this moment, the helpful types of de novo proteins are attracting an increasing amount of scientific energy and expertise, and the molecules may be coming to a clinic near you. As most of the world’s nearly eight billion people await a COVID vaccine, Walls’s nanoparticle is looking like a promising candidate.
After successfully neutralizing the pseudovirus in mouse cells, the vaccine’s next big test was against the real coronavirus. For that, Walls had to ship her mice to the University of North Carolina lab of Ralph S. Baric, one of the world’s foremost coronavirus researchers. The facility has the biosecurity level required to work with the live virus. Baric and his colleagues see many vaccine candidates, so in June 2020 Walls was pleased to get an encouraging e-mail from them: the neutralizing power of the nanoparticle vaccine was off the charts—higher than anything they had tested.
“Everything worked better than we’d hoped!” Walls says. When exposed to the real virus, the mice did well. “Completely protected. No sign of illness.” (Later Walls found that she could reduce the already low dose an additional ninefold, add a booster and get equally good results.) In January of this year the vaccine began early clinical trials in Washington State and South Korea.
Yet even as those trials were progressing, the virus was spawning a new wave of variants with the ability to evade some of the antibodies triggered by the first generation of vaccines. So Walls went back to work, designing a new and improved nanoparticle. Instead of copies of just the SARS-CoV-2 RBD, this version had a mosaic of four different RBDs: some from SARS-CoV-2, some from the original SARS virus from the early 2000s and some from two other coronaviruses. This broad spectrum of RBDs elicited a robust antibody response against all coronaviruses tested, including the most elusive of the variants.
A vaccine that is effective in tiny doses, that is easy and inexpensive to produce, that does not require refrigeration and that protects against a bunch of mutant viruses, including ones that may emerge in the future, could be exactly the solution the world needs. These advantages have drawn the attention of the world’s vaccine heavyweights, including GSK’s Rappuoli. “There is no question that our immune system likes nanoparticles,” he says. “These represent the best option we have.” In a recent commentary in the journal Cell, Rappuoli predicted that such designer molecules will usher in a new era of vaccines: “From here, the sky is the limit.”
And the capability will not end with vaccines. In this new Amino Age, the ability to intelligently design nanomachines at an atomic scale could turn fighting every disease into an engineering exercise. “When we tackle problems involving any sort of protein, we need to have this in mind,” Walls says. “We need to look at the protein and know that we can engineer solutions. Every day there are new successes coming.”
Some of those successes will come in areas other than medicine, such as materials science. The IPD has invented proteins that self-assemble into microscopic honeycomb grids that attract mineral deposition, a new way to produce efficient superconductors and batteries. Another project is crafting proteins that harvest light, as do photosynthetic proteins in plants, and convert that energy into electricity and fuel.
As the Amino Age tool kit grows, the natural proteins we now use for help—insulin for people with diabetes, for instance—may come to seem as archaic as the sharpened rocks our Stone Age ancestors once used. By the same token, our current designer proteins, as exciting as they are, are just sundials and wagon wheels. The features of a future landscape filled with bespoke molecules are beyond conception. But like the new proteins themselves, those features will, eventually and elegantly, fold into shape.
No comments:
Post a Comment